Computing
A quantum chemistry package.
The Department of Chemistry only makes this software available to research groups who have contributed towards the cost of acquiring the media.
Linux machines in Chemistry
Within the Department of Chemistry this package is installed on all managed Linux workstations but can only be used by research groups who have contributed to the cost of the software. Access is controlled by membership of the gaussian16 Unix group. If you are a member of the Department of Chemistry and your group would like to get access to the Linux software please email support@ch.cam.ac.uk for the price, which is per-group and covers as many machines as you want provided they are physically located at the University.
Members of groups who have contributed to the cost of the Linux software may also install it on unmanaged Linux machines physically located within the University. Please see here for details.
Gaussian 16 in other parts of the University
The licence Chemistry has arranged for Gaussian 16 covers the whole University. Other departments can access the software either by purchasing media sets directly from Gaussian, Inc or by arranging with Chemistry to pay a share of the licence cost and then getting the software via Chemistry. Contact support@ch.cam.ac.uk for details.
Other software from Gaussian
The University has site licences for Gaussian 09 for Linux and Mac, Gaussian 03 for Linux, Gaussview 5 for Linux and Mac, and Gaussview 6 for Linux and Mac.
On managed Linux machines load the gaussian16 module to access the software. The program itself is called g16. We have several different gaussian16 module versions available which support different CPU types. If you have trouble with Gaussian crashing with messages like ‘illegal instruction’ you probably need to try a module for a different CPU type. The gaussian16/16-A03/x86_64 module should work on all managed Linux machines but will not give the fastest performance on modern CPUs.
We also have Gaussview 6 (a graphical frontend to Gaussian) available. Loading the gaussian module will also make gaussview available in your environment. Type gview to start it.
New Chemistry with Gaussian 16 & GaussView 6
Continuing the nearly 40-year tradition of the Gaussian series of electronic structure programs, Gaussian 16 offers new methods and capabilities which allow you to study ever larger molecular systems and additional areas of chemistry. GaussView 6 offers a rich set of building and visualization capabilities. We highlight some of the most important features on this page.
Explore New Substances and Environments: Green Fluorescent Protein
GFP is a protein that fluoresces bright green when exposed to light in the blue-to-ultraviolet range. The chromophore is shown in the inset below. The molecule was first isolated in the jellyfish species Aequorea victoria , which is native to the Pacific northwest coast of North America. Since then, it has been studied extensively, and variants of the molecule with enhanced fluorescence properties have been engineered.
GFP consists of a chromophore within a protein chain composed of 238 amino acids. The isolated chromophore is not fluorescent, so modeling it in its protein environment is essential. GFP’s fluorescence cycle involves an initial excitation to its first excited state, a proton transfer reaction on the S1 potential energy surface, and finally a relaxation back to the ground state.
The following features of Gaussian 16 and GaussView 6 are useful for modeling fluorescence in this compound:
 |
|
- Gaussian can optimize the geometries of the minima and transition structures on the excited state PES with TD-DFT. GaussView includes features for setting up reliable QST2/QST3 transition structure optimizations with minimum effort.
- An IRC calculation in Gaussian can follow the corresponding S1 PES reaction path, which can then be animated in GaussView.
- Gaussian can perform vibrational frequency analysis in order to predict the IR/Raman spectra and normal modes. A variety of other spectra are also available, including vibronic spectra. GaussView can display plots of the predicted spectra and animate the associated normal modes (as applicable).
- GaussView makes it easy to examine the results of one calculation and then set up and initiate the next calculation in sequence via an intuitive interface to all major Gaussian 16 features.
Overview of What’s New in Gaussian 16
Gaussian 16 brings a variety of new methods, property predictions and performance enhancements. Details about many of them are given elsewhere in this brochure.
Modeling Excited States
- Analytic frequency calculations for the time-dependent (TD) Hartree-Fock and DFT methods, including ONIOM electronic embedding fully coupled with the environment of the MM region, without additional approximations [in prep.» target=»_blank»>WilliamsYoung17p].
- Geometry optimizations with the high accuracy EOM-CCSD method (analytic gradients) [J. Chem. Theory and Comput., 8 (2012) 5081-9. DOI: 10.1021/ct300382a» target=»_blank»>Caricato12a].
- Anharmonic analysis for calculating IR, Raman, VCD and ROA spectra [J. Chem. Phys.136 (2012) 124108. DOI: 10.1063/1.3695210″ target=»_blank»>Bloino12, JCTC8 (2012) 1015-1036. DOI: 10.1021/ct200814m» target=»_blank»>Bloino12a, J. Phys. Chem. A, 2015, 119, 11862–11874. DOI: 10.1021/acs.jpca.5b10067′ target=»_blank»>Bloino15]. Calculations in solution take the interaction between the excitation and the solvent field fully into account [J. Chem. Phys, 2011, 135, 104505. DOI: 10.1063/1.3630920′ target=»_blank»>Cappelli11].
- Vibronic spectra prediction [J. Chem. Theory Comput., 5 (2009) 540-54. DOI: 10.1021/ct8004744″ target=»_blank»>Barone09, Journal of Chemical Theory and Computation, 2010, 6, 1256-1274. DOI: 10.1021/ct9006772′ target=»_blank»>Bloino10, J. Chem. Theory Comp., 2013, 9, 4097-4115. DOI: 10.1021/ct400450k’ target=»_blank»>Baiardi13].
- Chiral spectroscopies: electronic circular dichroism (ECD) and circularly polarized luminiscence (CPL) [Phys. Chem. Chem. Phys . 14 (2012) 12404 — 422. DOI: 10.1039/C2CP41006K» target=»_blank»>Barone12, Chirality, 2014, 26, 588–600. DOI: 10.1002/chir.22325′ target=»_blank»>Barone14].
- Modeling of resonance Raman spectroscopy [Journal of Chemical Theory and Computation, 2014, 10, 346–363. DOI: 10.1021/ct400932e’ target=»_blank»>Egidi14, J. Chem. Phys., 2014, 141, 114108. DOI: 10.1063/1.4895534′ target=»_blank»>Baiardi14].
- Computation of electronic energy transfer (EET) [The Journal of Chemical Physics, 2004, 120, 7029. DOI: 10.1063/1.1669389′ target=»_blank»>Iozzi04].
- Ciofini’s excited state charge transfer diagnostic ( D CT) [J. Chem. Theory Comput., 2011, 7, 2498–2506. DOI: 10.1021/ct200308m ‘ target=»_blank»>LeBahers11, Coordination Chemistry Reviews, 2015, 304–305, 166–178. DOI: 10.1016/j.ccr.2015.03.027′ target=»_blank»>Adamo15].
- EOM-CCSD solvation interaction models of Caricato [J. Chem. Theory & Comput., 8 (2012) 4494. DOI: 10.1021/ct3006997″ target=»_blank»>Caricato12b].
New Methods
- Many DFT functionals have been added to Gaussian since the initial release of G09, including APFD [ J. Chem. Theory and Comput. 8 (2012) 4989. DOI: 10.1021/ct300778e» target=»_blank»>Austin12], functionals from the Truhlar group (most recently MN15 and MN15L [Chemical Science 2016, 7, 5032-5051. DOI: 10.1039/C6SC00705H.» target=»_blank» >Yu16]) and PW6B95 & PW6B95D3 [J. Phys. Chem. A, 2005, 109, 5656. DOI: 10.1021/jp050536c.’ target=»_blank»>Zhao05a].
- Additional double-hybrid methods: DSDPBEP86 [Phys. Chem. Chem. Phys., 2011, 13, 20104–20107, DOI: 10.1039/C1CP22592H’ target=»_blank»>Kozuch11], PBE0DH, PBEQIDH [The Journal of Chemical Physics, 2011, 135, 024106. DOI: 10.1063/1.3604569′ target=»_blank»>Bremond11,J. Chem. Phys., 2014, 141, 031101. DOI: 10.1063/1.4890314′ target=»_blank»>Bremond14].
- Empirical dispersion for a variety of functionals, using the schemes of Grimme (GD2, GD3, GD3BJ) [J. Comp. Chem., 27 (2006) 1787-99. DOI: 10.1002/jcc.20495″ target=»_blank»>Grimme06, J. Chem. Phys., 132 (2010) 154104. DOI: 10.1063/1.3382344″ target=»_blank»>Grimme10, J. Comp. Chem. 32 (2011) 1456-65. DOI: 10.1002/jcc.21759″ target=»_blank»>Grimme11] and others.
- The PM7 semi-empirical method, both in the original formulation [J. Molec. Modeling19 (2013) 1-32. DOI: 10.1007/s00894-012-1667-x’ target=»_blank»>Stewart13], and with modifications for continuous potential energy surfaces [in prep.» target=»_blank»>Throssel17p].
Performance Enhancements
Ease of Use Features
- Automatically recalculate the force constants every n th step of a geometry optimization.
- An expanded set of Link 0 commands and corresponding Default.Route file directives.
- Tools for interfacing Gaussian with external programs in compiled languages such as Fortran and C and/or in interpreted languages such as Python and Perl.
- Generalized internal coordinates: define arbitrary redundant internal coordinates and coordinate expressions for use as geometry optimization constraints.
Overview of What’s New in GaussView 6
GaussView 6 provides support for all major Gaussian 16 features as well as offering additional modeling capabilities of its own.
Building and Manipulating Molecular Structures
- Select step(s) to open from multi-job Gaussian input files.
- New advanced open dialog, allowing options to be modified and retained across sessions: whether to load intermediate geometries, weak bond inclusion, PDB file processing options, and similar settings.
- New brush selection mode to accurately choose atoms within dense regions.
- Open PDB files created by Amber.
- Reduce symmetry of a molecule to a specified point group (using the same feature as for increasing and constraining symmetry).
- Specify custom bonding parameters.
Gaussian Job Setup & Execution
- Support for new features in Gaussian 16 and updates to existing job setup options.
- Create and/or initiate identical calculations for a series of molecules in a single step.
- Create new Gaussian input files which use the molecule specification and other data from a checkpoint file.
- Support for a wide range of Link 0 commands.
- Expanded access to population analysis options.
- Preview the input file as it is created.
- The SC Job Manager provides a straightforward, customizable queueing system for the local computer.
Additional Modeling Capabilities
- Perform conformational searches using the GMMX add-on package. GaussView allows you to:
- Specify the force field, energy range and other parameters for the search.
- Open some or all of the identified conformations into a molecule group.
- View an energy plot for the set of conformations.
- Set up follow-up calculations for desired conformations in a single step.
- Optional full AMPAC interface integration.
Examine & Visualize Results
- Select step(s) to open from multi-calculation Gaussian results files.
- Visualize predicted bond lengths and bond orders.
- Results from anharmonic frequency calculations can be plotted alone or in combination with the harmonic results.
- Include multiple calculation results in the same plot. Spectra from multiple conformations can be Boltzmann-averaged.
- Customize plots: specify line colors and widths, canvas and background colors, label and title fonts, X- and Y-axis settings, and so on.
- Display the solvation cavity surface for an SCRF calculation.
- Plot the results of Optical Rotary Dispersion (ORD) calculations.
- Scale frequencies by a specified value for plotting.
- Support for Gaussian trajectory calculations.
- Image capture enhancements: background color, save images of multiple molecules in a single operation.
- Image capture and printing defaults.
- Enhanced movie creation: support for MPEG4 files; set options for frame looping and time delays.
File Conversion & Customization
- Convert files between different formats; AMPAC input files, Gaussian input files, MDL Mol and SDF files, GMMX input files and Sybyl Mol2 files.
- Set defaults for image capture, movie generation and custom bonding parameters.
- Context-specific help system.
Sophisticated Modeling of Excited States
Excited state methods and properties received a lot of attention as we developed Gaussian 16. With its analytic TD-DFT frequencies, you can optimize excited state transition structures and perform IRC calculations. Analytic EOM-CCSD gradients let you optimize the structures of molecules that require a high accuracy excited state treatment.
Anharmonic Vibrational Analysis
Standard vibrational frequency analysis employs the double harmonic approximation, the simplest description of the vibrations and the molecule’s interaction with the electromagnetic field of the incident photons. It treats nuclear vibrations as simple harmonic motion about the minimum energy structure and uses a correspondingly simplified model for the electromagnetic field interaction. For many systems, this is sufficient, but sometimes the approximation reduces the accuracy of the predicted spectra.
Anharmonic frequency analysis relaxes both parts of the double harmonic approximation by introducing additional mathematical terms: higher derivatives of the energy, dipole moment, polarizability (as appropriate to the type of spectroscopy being modeled). Doing so results in changes in the locations of the fundamental frequencies and enables the prediction of overtone and combination bands. Later revisions of Gaussian 09 included anharmonic IR and Raman spectra, and Gaussian 16 adds anharmonic VCD and ROA spectra.
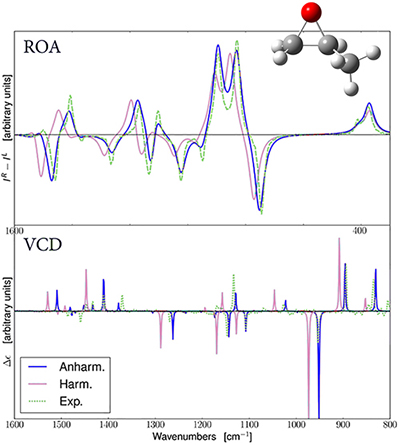
Predicted ROA and VCD Spectra for R-Methyloxirane
Resonance Raman Spectroscopy
Gaussian 16 can predict Resonance Raman (RR) spectra. RR spectroscopy is a type of Raman spectroscopy where the incident laser frequency is close in energy to an electronic transition of the compound being studied. This has the advantage of enhancing the intensity of the scattered light.
These experiments typically scan over a range of frequencies for the incident light. The resulting data can be plotted as a three dimensional surface: the incident light frequencies and the corresponding observed peaks are plotted in the X-Y plane, with the intensity determining the height of the curve above the plane.
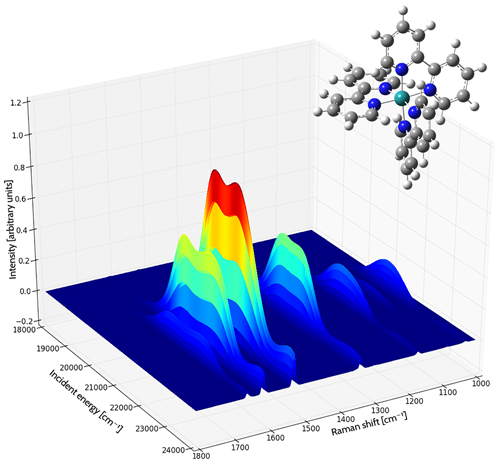
Resonance Raman Spectrum of the tris(2,2’-bipyridyl)-ruthenium(II) Complex
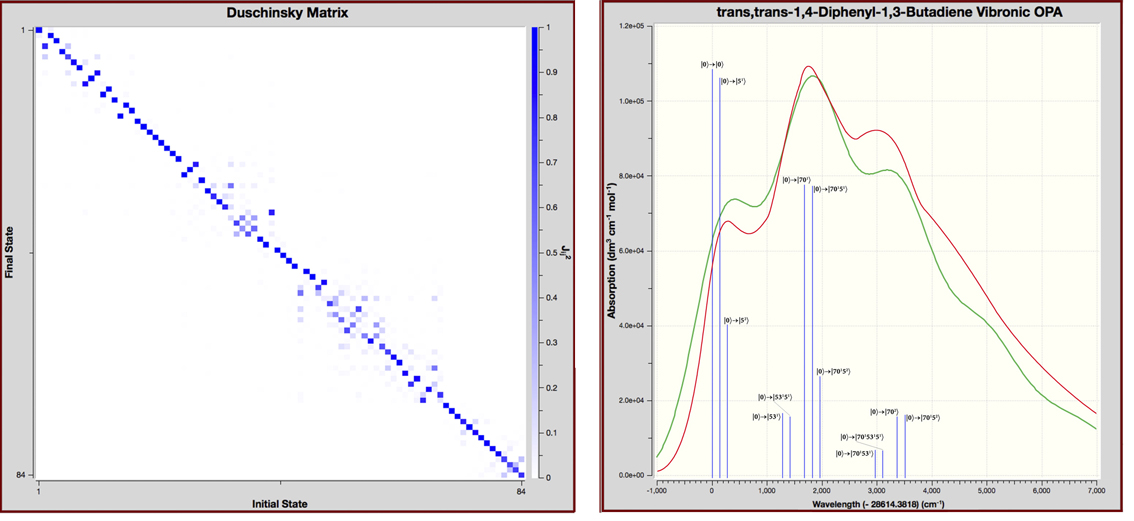
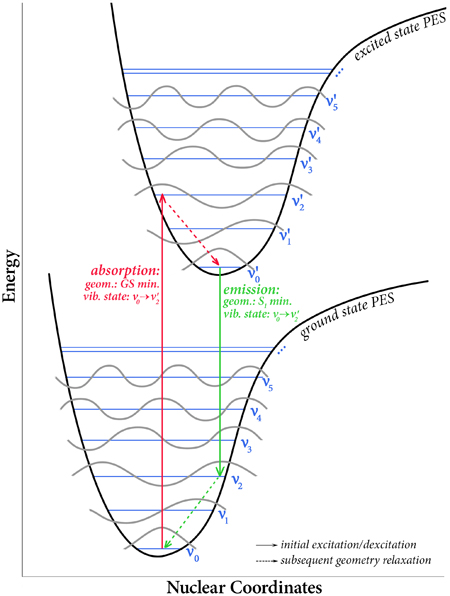
Franck-Condon/Herzberg-Teller Analysis and Vibronic Spectra
Gaussian 16 can compute vibronically-resolved electronic spectroscopy for one photon absorption processes. In a vertical excitation resulting from the absorption of a photon, a molecule moves from its ground electronic state to an excited state. The geometry of the excited state immediately afterwards is not the equilibrium structure of the excited state, i.e., the minimum on the excited state potential energy surface. Rather, because the time scale in which electronic transitions occur is so small compared to nuclear motions, the nuclei are nearly unaffected, and the ground state equilibrium geometry is initially retained.
 |
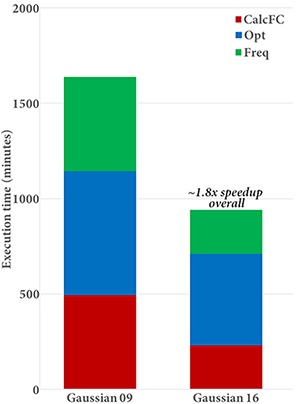 | 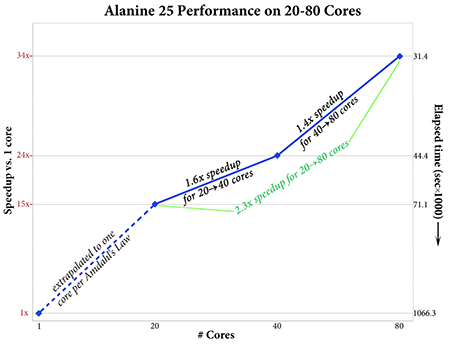 |
Gaussian 16’s performance scales effectively to 80 cores or more (depending on the calculation and molecule). The plot on the right above gives the speedups for 20, 40 and 80 cores for an APFD/6-31G(d) Freq calculation on Alanine 25. This job shows good parallel speedups with increasing numbers of processors; the speedup when moving from 20 to 80 cores is about 2.3, and the estimated speedup for 80 cores over a single processor is 34.
Algorithmic Improvements
Gaussian 16 also offers a wide variety of other performance improvements to many different parts of the program. Among the most significant are the following:
Running Calculations on GPUs with Gaussian 16
As a result of a fruitful, ongoing collaboration between the Gaussian Inc., NVIDIA and its PGI compiler team and Hewlett-Packard Enterprise, Gaussian 16 supports running calculations using NVIDIA GPUs. NVIDIA Tesla K40, Tesla K80, Tesla P100 and Tesla V100 GPUs can be used in Hartree-Fock and DFT calculations, including energies, optimizations and frequencies, for ground and excited states (TD), and for closed shell and open shell molecules. ONIOM, SCRF solvation and all major properties are supported, as are all DFT functionals available in Gaussian 16, making the most frequently-run Gaussian calculations applicable to execution with GPUs.
NVIDIA Tesla V100 GPUs use the NVIDIA Volta GPU architecture to achieve
7 (PCIe) TFlops peak performance (double precision), and have 16GB-32GB HBM2 memory.
NVIDIA Tesla P100 GPUs use the NVIDIA Pascal GPU architecture to achieve
5 TFlops peak performance (double precision), and have 12-16GB HBM2 memory.
NVIDIA Tesla K40 & Tesla K80 GPUs have 12GB 5GHz GDDR5 VRAM and achieve peak performance of
1.5 TFlops (double precision), with 1 and 2 GPUs per board (respectively).
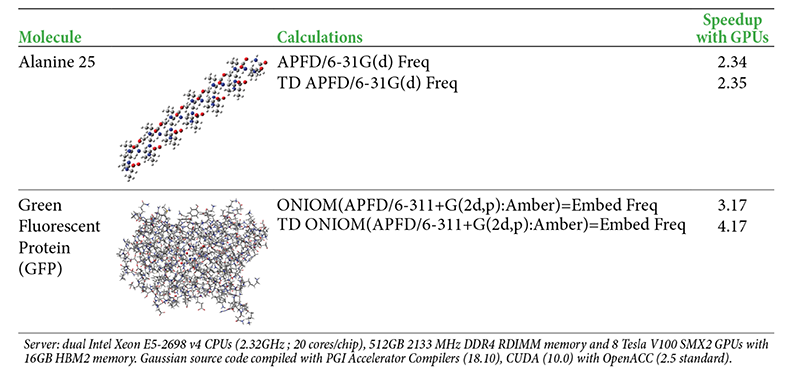
Performance Results for Example Calculations
The following table provides some example performance data for the current version of Nvidia V100 GPU support. The timings compare running a calculation across 28 CPU cores on a DGX-1 server (Broadwell CPUs) versus running on the same server across 28 CPU cores and 8 GPUs (with 8 CPU cores serving as GPU controllers during the GPU-enabled parts of the calculation).
GPU Parallelization Strategy
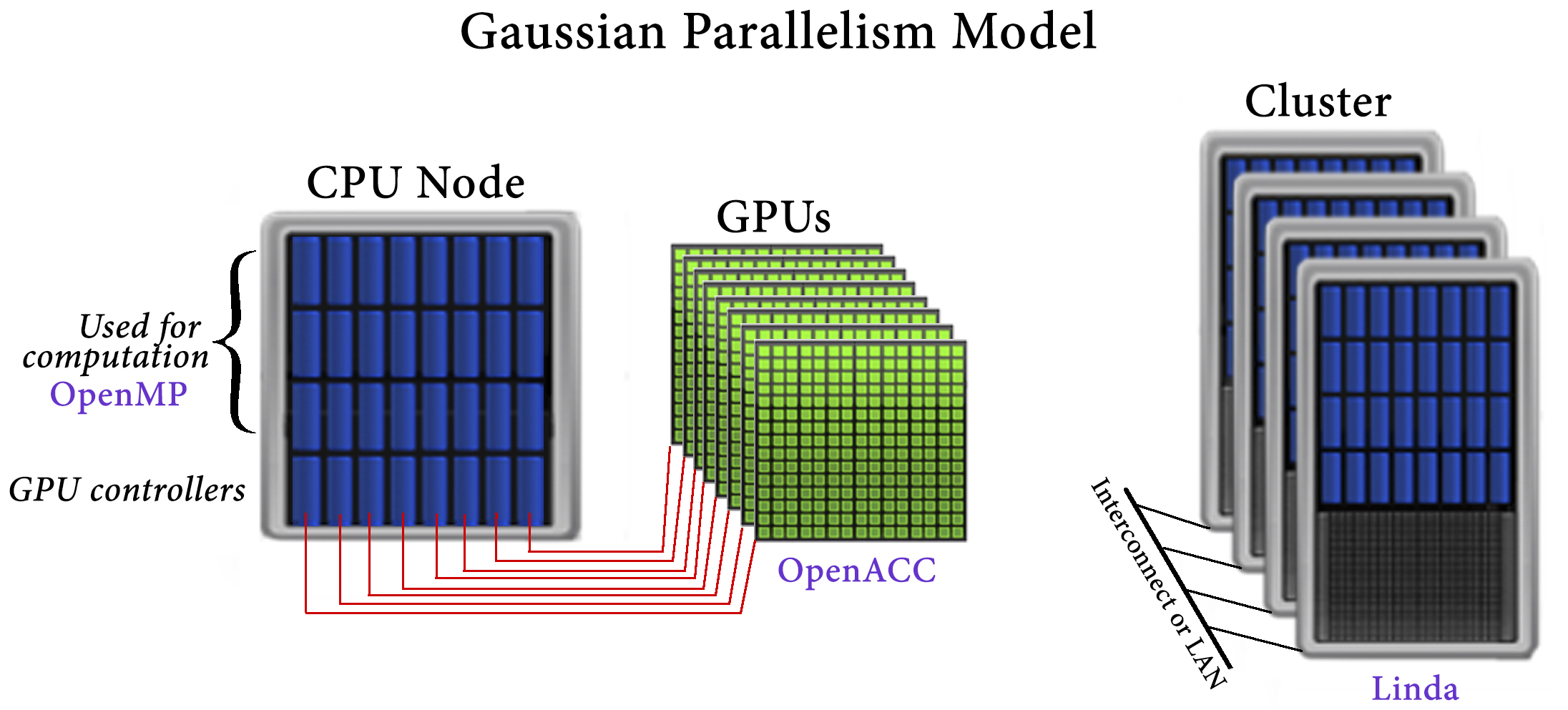
Within Gaussian 16, GPUs are used for a small fraction of code that consumes a large fraction of the execution time. The implementation of GPU parallelism conforms to Gaussian’s general parallelization strategy. Its main tenets are to avoid changing the underlying source code and to avoid modifications which negatively affect CPU performance. For these reasons, OpenACC was used for GPU parallelization.
The Gaussian approach to parallelization relies on environment-specific parallelization frameworks and tools: OpenMP for shared-memory, Linda for cluster and network parallelization across discrete nodes, and OpenACC for GPUs.
The process of implementing GPU support involved many different aspects:
- Identifying places where GPUs could be beneficial. These are a subset of areas which are parallelized for other execution contexts because using GPUs requires fine grained parallelism.
- Understanding and optimizing data movement/storage at a high level to maximize GPU efficiency.
- PGI’s sophisticated profiling and performance evaluation tools were vital to the success of the effort.
PGI Accelerator Compilers with OpenACC
PGI compilers fully support the current OpenACC standard as well as important extensions to it. PGI is an important contributor to the ongoing development of OpenACC.
OpenACC enables developers to implement GPU parallelism by adding compiler directives to their source code, often eliminating the need for rewriting or restructuring. For example, the following Fortran compiler directive identifies a loop which the compiler should parallelize:
Other directives allocate GPU memory, copy data to/from GPUs, specify data to remain on the GPU, combine or split loops and other code sections, and generally provide hints for optimal work distribution management, and more.
The OpenACC project is very active, and the specifications and tools are changing fairly rapidly. This has been true throughout the lifetime of this project. Indeed, one of its major challenges has been using OpenACC in the midst of its development. The talented people at PGI were instrumental in addressing issues that arose in one of the very first uses of OpenACC for a large commercial software package.